Michael G. Lerner
Research: Bilayers
Note: this page is currently being updated. For more recent papers on bilayers and diffusion, see my Google Scholar Profile.System-size effects
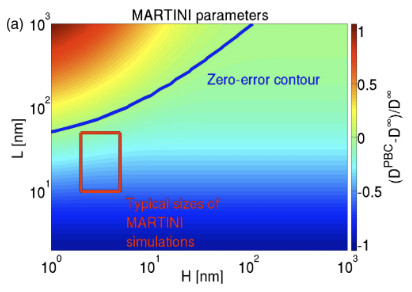
Molecular dynamics simulations are typically carried out in periodic boundary conditions (PBC). Those PBC can have dramatic impacts on dynamic properties, like membrane diffusion. Yeh and Hummer showed that the size of a simulated box of water molecules can have moderate effects on the diffusion constants seen in those systems. We used theory and simulation to show (1, 2) that, in simulations of membranes, the system size has a very large effect on the observed diffusion. We are currently working to extend these models to look at correlated motions in simulated systems.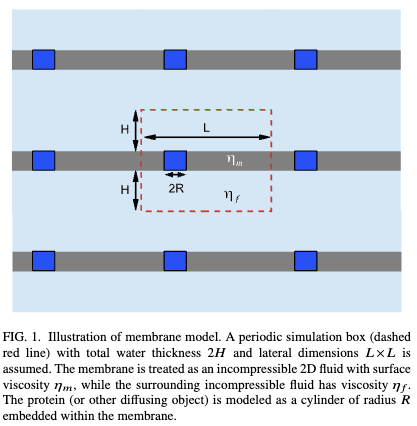
Diffusion of membrane-bound dimer and trimer systems
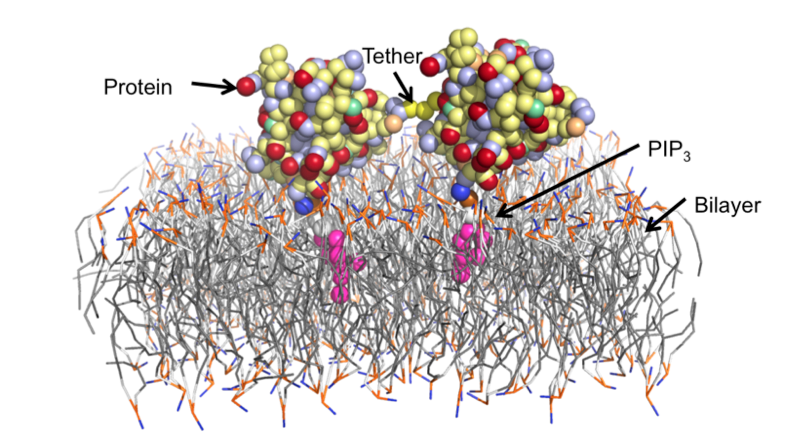
Membrane-targeting proteins are recruited to specific membrane surfaces during cell signaling events. These interactions frequently involve the recruitment of multiple lipids including phosphatidylserine (PS), phosphatidylinositol-4,5-bisphosphate (PIP2), phosphatidylinositol-3,4,5-trisphosphate (PIP3) and diacylglycerol (DAG). Interactions with proteins are highly regulated. The pleckstrin homology (PH) domain specifically binds PIP3, and several membrane-targeting proteins have multiple PH domains. As a more complex example, protein kinase C has three specific binding sites, one for each of PS, PIP3 and DAG, and all three are required for full activation. Determining the number and types of bound lipids is thus crucial to understanding the activity of membrane-targeting proteins.
We have developed computational and theoretical models to determine a precise and readily measurable relationship between the lateral diffusion constant of a protein and the number of specifically bound lipids[1,2]. The correspondence with experimental work done in collaboration with Joe Falke and Jeff Knight is excellent. The computational models involve the Martini force field, and are simulations are run in GROMACS and CHARMM. The analytical results are based primarily on the Saffman-Delbruck model of a cell membrane, using the formalism of velocity response functions.
This work has been highlighted in the Biophysical Society newsletter, and was the second most read Biophysical Journal paper in all of July-December 2010.
We we worked to extend the model to include larger systems, resulting in the series of papers on system-size effects described above.
-
Single molecule diffusion of membrane-bound proteins: Window into lipid contacts and bilayer dynamics
Jefferson D. Knight, Michael G. Lerner, Joan G. Marcano-Velazquez, Richard W. Pastor and Joseph J. Falke
Biophysical Journal 99(9):2870-2887 (2010) [pdf | online]